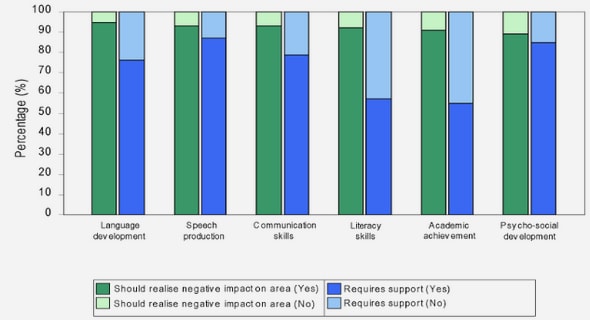
(Downloads - 0)
For more info about our services contact : help@bestpfe.com
Table of contents
General introduction
Quantitative genetics in the genomic era
Genetic structure: theory, inference and a maize perspective
How does genetic structure affect quantitative traits?
Impact of genetic structure on association mapping and genomic selection
Objectives of the thesis
1 Genomic selection efficiency and a priori estimation of accuracy in a structured dent maize panel
Abstract
Introduction
Materials and methods
Genetic material and genotypic data
Structure analysis
Phenotypic data
Genomic prediction models
Evaluation of the precision of genomic predictions
A priori estimation of accuracy
Results
Global, within and across group precision of genomic predictions
Accounting for structure in genomic prediction models
A priori estimation of precision
Discussion
The impact of genetic structure on genomic prediction accuracy
Modeling genetic structure to improve predictions
Is it possible to forecast accuracy using CD?
Conclusion
2 Disentangling group specific QTL allele effects from genetic background epistasis using admixed individuals in GWAS: an application to maize flowering
Abstract
Introduction
Materials and methods
Genetic material and genotypic data
Phenotypic data
Global assessment of directional epistasis
GWAS models
Results
Phenotypic analysis and directional epistasis
Associations detected and GWAS strategies
Highlighted QTLs
Discussion
Accounting for genetic groups in GWAS
Benefits from admixed individuals
Heterogeneity of maize flowering QTL allele effects
Conclusion
Appendix A
3 Accounting for group-specific allele effects and admixture in genomic predictions: theory and experimental evaluation in maize
Abstract
Introduction
Statistical context
Statistical context
MAGBLUP
Material and Methods
Flint-Dent dataset
Statistical inference and genomic predictions
Simulated traits
Assessment of the precision of variances estimates
Assessment of the accuracy of genomic predictions
Results
Variance estimates for simulated traits
Genomic prediction accuracy for simulated traits
Application to real traits
Discussion
Modeling group-specific allele in admixed populations
Variance components and genomic predictions
Benefits from admixed individuals in multi-group training sets
Conclusion
Appendix A
Appendix B
Appendix C
Appendix D
Appendix E
Appendix F
Appendix G
Appendix H
General discussion
Perspectives
Bibliography