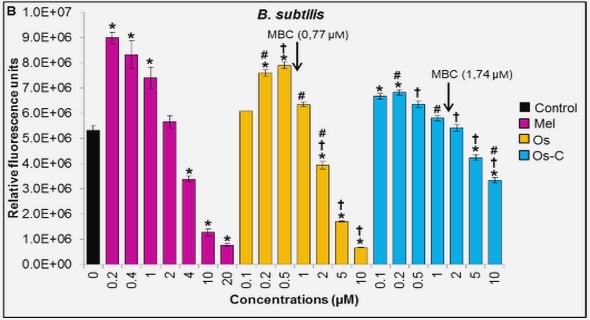
(Downloads - 0)
For more info about our services contact : help@bestpfe.com
Table of contents
1 Chapter 1 Background
1.1 The history of acetone-butanol-ethanol (ABE) fermentation
1.2 Clostridium acetobutylicum strain
1.3 The central metabolism of Clostridium acetobutylicum
1.3.1 Glycolysis
1.3.2 Enzymes involved in electron fluxes
1.3.3 Central pathway from acetyl-CoA to butyryl-CoA
1.3.4 Acetate formation
1.3.5 Butyrate formation
1.3.6 Lactate and acetoin formation
1.3.7 Acetone formation
1.3.8 Ethanol and butanol formation
1.4 Metabolic engineering tools in Clostridium acetobutylicum
1.4.1 Shuttle vectors for gene over-expression
1.4.2 Inducible promoter/repressor systems
1.4.3 Reporter systems
1.4.4 Antisense RNA (asRNA)
1.4.5 Group II intron
1.4.6 Genomic mutagenesis by homologous recombination system
1.4.7 Random mutagenesis by mariner transposon system
1.5 Clostridium acetobutylicum as an n-butanol producer-a challenging task
1.6 Genome scale metabolic (GSM) models for Clostridium acetobutylicum
1.7 Regulatory network in Clostridium acetobutylicum
1.7.1 Spo0A and Sigma factors
1.7.2 The controversial SolR
1.7.3 Agr quorum sensing
1.7.4 Heat-shock proteins: DnaK, GroESL, and Hsp21
1.7.5 PhoPR
1.7.6 AbrB310
1.7.7 Per and Fur
1.7.8 CsrA
1.7.9 CcpA
1.7.10 Rex
1.7.11 AraR
1.7.12 Small regulatory RNA
2 Chapter 2 Thesis Objectives
3 Chapter 3 Construction of a restriction-less, marker-less mutant useful for functional genomic and metabolic engineering of the biofuel producer Clostridium acetobutylicum
3.1 Abstract
3.2 Introduction
3.3 Results and Discussion
3.3.1 MGCΔcac1502 strain, a C. acetobutylicum strain that is transformable without previous in vivo plasmid methylation
3.3.2 Construction of the MGCΔcac1502Δupp strain: the first marker-free C. acetobutylicum strain with two deleted genes
3.3.3 Deletion of the CA_C3535 gene in the MGCΔcac1502Δupp strain using the upp/5-FU system as a counter-selectable marker for the loss of plasmid
3.3.4 Determination of the recognition sequence of Cac824II encoded by CA_C3535
3.3.5 Deletion of the ctfAB genes in the MGCΔcac1502ΔuppΔcac3535 to create a strain no longer producing acetone
3.3.6 Deletion of the ldhA gene in the MGCΔcac1502ΔuppΔcac3535 to create a strain no longer producing lactate
3.4 Discussion
3.5 Conclusion
3.6 Experimental Procedure
3.6.1 Bacterial strain, plasmids and oligonucleotides
3.6.2 Culture and growth conditions
3.6.3 DNA manipulation techniques
3.6.4 Construction of pUC18-FRT-MLS2
3.6.5 Construction of pCons2.1
3.6.6 Construction of pCIP2-1
3.6.7 Construction of pREPcac15
3.6.8 Construction of pCIPcac15
3.6.9 Construction of pREPupp
3.6.10 Construction of pCLF1
3.6.11 Construction of pCons::upp
3.6.12 Construction of pREPcac35::upp
3.6.13 Construction of pREPctfAB::upp
3.6.14 Construction of pREPldhA::upp
3.6.15 Construction of pCLF::upp
4 Chapter 4 The Weizmann process revisited for the continuous production of n-butanol
4.1 Abstract
4.2 Main text
4.3 Supplementary Materials and Methods
4.3.1 Bacterial strains, plasmids and primers
4.3.2 Chemostat culture of recombinant strains
4.3.3 Continuous extractive high density cell recycle fermentation
4.3.4 Measurement of fermentation parameters
4.3.5 General protocol for gene deletion by homologous recombination system using replicative plasmid pSOS95-MLSr [167,174]
4.3.6 Construction of pSOS95-MLSr-upp-Drex-catP and CAB1058 strain
4.3.7 Construction of pEryupp-atoB plasmid and CAB1059 strain
4.3.8 Construction of pSOS95-upp-hbd1-catP-oriRepA and CAB1060 strain
4.3.9 Enzyme activity measurements
5 Chapter 5 Construction of a hydrogenase minus mutant of Clostridium acetobutylicum : a platform strain for the continuous production of fuels and chemicals
5.1 Main text
5.2 Conclusions
5.3 Material and Methods
5.3.1 Knockout of hydA and thlA using LltrB intron
5.3.2 Construction and integration by single crossing over of the pEryUppΔthlA plasmid in the chromosome of C. acetobutylicumuppcac1502
5.3.3 Construction and integration by single crossing over of the pCatUppHydA270 plasmid in the chromosome of C. acetobutylicumuppcac1502
5.3.4 Simultaneous inactivation of thlA and hydA in C. acetobutylicumuppcac1502 116
6 Chapter 6 Cap0037, a novel global regulator of Clostridium acetobutylicum metabolism
6.1 Abstract
6.2 Introduction
6.3 Results and Discussion
6.3.1 The phylogenetic tree and bioinformatics analysis of Cap0037/Cap0036
6.3.2 Disruption of CA_P0037 by pCUI-cap37 (189s)
6.3.3 Carbon and electron fluxes of the Cap37::intron mutant under different physiological conditions
6.3.4 Determination of the CA_P0037 DNA binding site (BS)
6.3.5 Determination of putative Cap0037 regulon
6.3.6 Global transcription changes in the Cap37::int mutant
6.4 Conclusion
6.5 Experimental Procedure
6.5.1 Bacterial strains, plasmids and culture media
6.5.2 Plasmid construction
6.5.3 Continuous culture
6.5.4 Isolation of total mRNA and microarray
6.5.5 Analytical methods
6.5.6 Southern blot analysis
6.5.7 Expression and purification of His-tagged protein
6.5.8 Electromobility shift assays (EMSAs)
6.5.9 DNase I protection assays (DNA Footprinting)
6.5.10 Bioinformatic tools
6.5.11 Microarray data accession number
6.6 Acknowlegdement
6.7 Supplementary
7 Chapter 7 Conclusion and future perspectives